About¶
Network-level analysis of various features, esp. if it can be individualized for a single-subject, is proving to be quite a valuable tool in many applications. This package extracts single-subject (individualized, or intrinsic) networks from node-wise data by computing the edge weights based on histogram distance between the distributions of values within each node. Individual nodes could be an ROI or a patch or a cube, or any other unit of relevance in your application. This is a great way to take advantage of the full distribution of values available within each node, relative to the simpler use of averages (or another summary statistic).
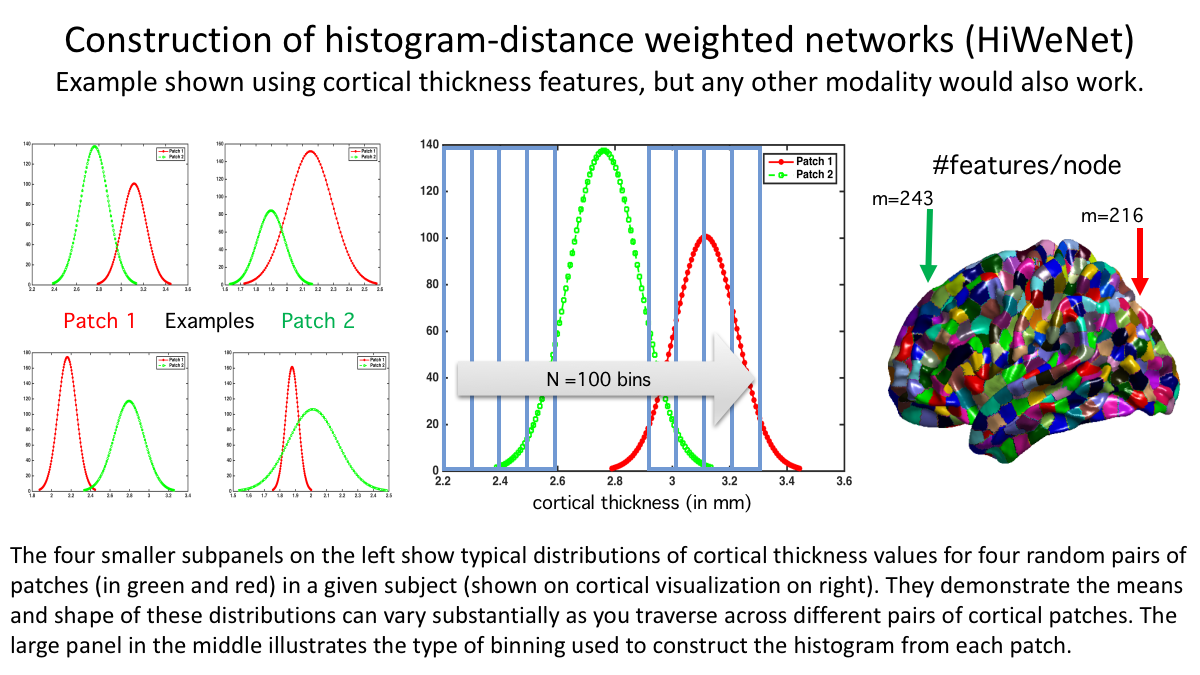
Rough scheme of computation is shown below:
Applicability¶
Although this technique was originally developed for cortical thickness, this is a generic and powerful technique that could be applied to any features such as gray matter density, PET uptake values, functional activation data or EEG features. All that is needed is a set of nodes/parcellation that have one-to-one correspondence across samples/subjects in your dataset.
The target audience is users of almost all neuroimaging modalities who:
preprocessed dataset already,
have some base features extracted (node- or patch-wise, that are native to the given modality) using other packages (metioned above), and
who would like to analyze network-level (i.e. covariance-type or connectivity) relations among the base features (either in space across the cortex or a relevant domain, or across time).
This is similar to popular metrics of covariance like Correlation or LedoitWolf, and could be dropped in their place. Do you want to find out how histogram-based method compare to your own ideas?
What does hiwenet do?¶
This packages takes in vector of features and their membership labels (denoting which features belong to which groups - alternatively referred to as nodes in a graph), and computes their pair-wise histogram distances, using a chosen method.
This package is designed to be domain-agnostic, and hence a generic input format was chosen.
However, we plan to add interfaces to tools that may be of interest to researchers in specific domains such as nilearn, MNE and the related. A scikit-learn compatible API/interface is also in the works.
Refer to Examples using API and API Reference pages for more detailed and usage examples, and examples directory for sample files.
Thanks for checking out. Your feedback will be appreciated.
Citation¶
If you found this toolbox useful for your research, please cite one or more of these papers in that order:
Raamana, P. R. and Strother, S.C., 2017, Histogram-weighted Networks for Feature Extraction, Connectivity and Advanced Analysis in Neuroscience. Journal of Open Source Software, 2(19), 380, doi:10.21105/joss.00380
Raamana, P.R. and Strother, S.C., 2017, Impact of spatial scale and edge weight on predictive power of cortical thickness networks bioRxiv 170381 http://www.biorxiv.org/content/early/2017/07/31/170381.
Raamana, P. R., Weiner, M. W., Wang, L., Beg, M. F., & Alzheimer’s Disease Neuroimaging Initiative. (2015). Thickness network features for prognostic applications in dementia. Neurobiology of aging, 36, S91-S102.
Acknowledgements¶
I would like to thank Oscar Esteban (@oesteban) for his volunteer and attentive review of this package, which has been very helpful in improving the software.